Did you ever wonder how scientists genetically engineer organisms to express a new trait? Would you like to trace your ancestry genetically? Does forensic science and solving crimes fascinate you? AP Biology students got the opportunity to answer those questions with the help of three exciting laboratory experiences with Cold Spring Harbor’s DNA Learning Center.
On September 15th, three educators from the Dolan DNA Learning Center at Cold Spring Harbor visited the Lycée Français to assist AP Biology students in genetically engineering a strain of E. coli. Although this common type of bacteria can cause disease, this strain had been rendered harmless. Small circular pieces of DNA called plasmids containing a gene coding for green fluorescent protein and a gene coding for resistance to the antibiotic ampicillin, were inserted into the E. coli.
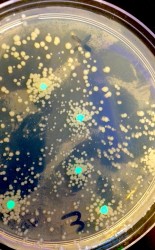
The photo on the right shows the result of an experiment conducted in the LFNY lab. Six E. coli colonies are seen expressing the green fluorescent protein and growing on a nutrient agar containing ampicillin. The bacteria are resistant to the antibiotic ampicillin.
After an incubation period of several days, the bacterial plates were examined for recombinant bacteria containing the new genes. Several colonies of bacteria, which had been successfully modified for resistance to ampicillin, grew on nutrient agar containing ampicillin and were glowing, under ultraviolet light, with the green fluorescent protein!
Tracing the Ancestry of Human Populations
Excited to develop their biotechnology skills, the AP Biology students traveled to Cold Spring Harbor’s Dolan DNA Learning Center on October 2nd and to the Harlem DNA Learning Center on October 15th. To carry out the experiments, each student obtained several thousand cheek cells from a saline mouthwash (bloodless and noninvasive).

A student amplifies a DNA sequence via a technique called polymerase, during a field trip at Cold Spring Harbor’s DNA lab.
At Cold Spring Harbor, a technique called polymerase chain reaction (PCR) was used to amplify a DNA sequence from chromosome 16 called PV92. It is a short nucleotide sequence called Alu within a noncoding region of the chromosome. The Alu family is a group of repeated DNA sequences found throughout primate genomes and can be used to analyze, in a limited way, the ancestry of human populations. Over the past 65 million years, the Alu sequence has amplified to about 1,000,000 copies, comprising an estimated 10% of the human genome. Many of these Alu insertions are only found in humans. Some of these are as recent as 1-2 million years ago in human populations.
Students screened their own DNA for the presence of the Alu insertion. By adding specific primers, which bind to the PV92 locus on chromosome 16 during PCR, students amplified their two versions of the Alu PV92 gene. Either they had two genes which were 715 base pairs long or two genes which were 415 base pairs long or one of each gene. By adding their DNA to a gel electrophoresis apparatus, students could determine their genotypes for the Alu PV92 gene. A great learning experience was had by all!
FBI Fingerprinting Method
At the Harlem DNA learning Center, students were introduced to The Federal Bureau of Investigation’s 13-marker panel of short DNA sequences used to create an individual’s DNA fingerprint for purposes of criminal investigations, determination of paternity or the identification of remains.

Dr. Bruce Nash, of the Cold Spring DNA Learning Center, shows Y11 and Y12 AP biology students how to isolate cells from their mouth using saline solution and amplify one region of their DNA using polymerase chain reaction. The students spent the entire day at the lab, located in Long Island, to learn about human genetics and polymorphisms.
Called Short Tandem Repeats (STR’s) or Variable Number Tandem Repeats (VLTR’s), these sequences do not code for any traits – not even the obvious ones like hair color or skin color. Instead, a DNA fingerprint identifies how many times these short sequences are repeated in each of the 13 locations. Everyone has these repeated sequences in the same locations but with different numbers of repeats. The probability of having the same number of repeats as another person of the same ethnic group is calculated for each of the 13 loci. When those probabilities are multiplied together, the chances of one person’s DNA fingerprint matching another person’s DNA fingerprint may be in the tens of billions – with the exception of identical twins. Thus, modern DNA testing has the capability to identify each person alive today.
This system is called the FBI’s Combined DNA Index System (CODIS). The students examined a VNTR on chromosome 1 called D1S80 (or pMCT118), which has a repeat unit of 16 base pairs. Most individuals have between 14 and 40 copies of the repeat on each copy of chromosome 1. After amplification through PCR, the student DNA samples were loaded onto a DNA chip and processed through a bioanalyzer. Students were given their individual genotypes for one of the FBI’s 13 markers. It was a very fun day!
About the Author :
Fran Moss retired from the Bronx High School of Science in June of 2010, after 26 years with the New York City Department of Education, and started teaching AP Biology at the Lycee the following September. She has a Master’s degree in Secondary Science Education from City College of the City University of New York. She was born in Trenton, New Jersey, and lives in Riverdale with her husband. She has two sons and two stepsons.